The Using DNA Barcodes to Identify and Classify Living Things laboratory demonstrates several important concepts of modern biology. During the course of this laboratory, you will:
- Collect and analyze sequence data from plants, fungi, or animals – or products made from them.
- Use DNA sequence to identify species.
- Explore relationships between species.
In addition, the laboratory experiment utilizes several experimental and bioinformatics methods in modern biological research. You will:
- Collect plants, animals, or products in your local environment or neighborhood.
- Extract and purify DNA from tissue or processed material.
- Amplify a specific region of the chloroplast, mitochondrial, or nuclear genome by polymerase chain reaction (PCR), and analyze PCR products by gel electrophoresis.
- Use the Basic Local Alignment Search Tool (BLAST) to identify sequences in databases.
- Use multiple sequence alignment and tree-building tools to analyze phylogenetic relationships.
Taxonomy, the science of classifying living things according to shared features, has always been a part of human society. Carl Linneas formalized biological classification with his system of binomial nomenclature that assigns each organism a genus and species name.
Identifying organisms has grown in importance as we monitor the biological effects of global climate change and attempt to preserve species diversity in the face of accelerating habitat destruction. We know very little about the diversity of plants and animals – let alone microbes – living in many unique ecosystems on earth. Less than two million of the estimated 5-50 million plant and animal species have been identified. Scientists agree that the yearly rate of extinction has increased from about one species per million to 100-1,000 per million. This means that thousands of plants and animals are lost each year. Most of these have not yet been identified.
Classical taxonomy falls short in this race to catalog biological diversity before it disappears. Specimens must be carefully collected and handled to preserve their distinguishing features. Differentiating subtle anatomical differences between closely related species requires the subjective judgment of a highly trained specialist – and few are being produced in colleges today.
Now, DNA barcodes allow non-experts to objectively identify species – even from small, damaged, or industrially processed material. Just as the unique pattern of bars in a universal product code (UPC) identifies each consumer product, a “DNA barcode” is a unique pattern of DNA sequence that can potentially identify each living thing. Short DNA barcodes, about 700 nucleotides in length, can be quickly processed from thousands of specimens and unambiguously analyzed by computer programs.
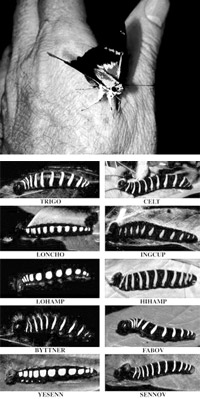
DNA barcoding revealed that what was once thought to be one species of butterfly is really ten species with caterpillars that eat different plants.
The International Barcode of Life (iBOL) organizes collaborators from more than 150 countries to participate in a variety of “campaigns” to census diversity among plant and animal groups – including ants, bees, butterflies, fish, birds, mammals, fungi, and flowering plants – and within ecosystems – including the seas, poles, rain forests, kelp forests, and coral reefs. The 10-year Census of Marine Life, completed in 2010, provided the first comprehensive list of more than 190,000 marine species and identified 6,000 potentially new species.
There is a surprising level of biological diversity, literally in front of our eyes. For example, DNA barcodes showed that a well-known skipper butterfly (Astraptes fulgerator), identified in 1775, is actually ten distinct species. DNA barcodes have revolutionized the classification of orchids, a complex and widespread plant family with an estimated 20,000 members. The urban environment is also unexpectedly diverse; DNA barcodes were used to catalogue 54 species of bees and 24 species of butterflies in community gardens in New York City.
DNA barcodes are also used to detect food fraud and products taken from conserved species. Working with researchers from Rockefeller University and the American Museum of Natural History, students from Trinity High School found that 25% of 60 seafood items purchased in grocery stores and restaurants in New York City were mislabeled as more expensive species. One mislabeled fish was the endangered species, Acadian redfish. Another group identified three protected whale species as the source of sushi sold in California and Korea. However, using DNA barcodes to identify potential biological contraband among products seized by customs is now well established.
Barcoding relies on short, highly variably regions of the genome. Although there is no universal barcode, a growing list of variable regions can help differentiate species from diverse taxonomic groups. With thousands of copies per cell, mitochondrial and chloroplast sequences are readily amplified by polymerase chain reaction, even from very small or degraded specimens. Regions of chloroplast genes, including rbcL (RuBisCo—Ribulose-1,5-bisphosphate carboxylase oxygenase—large subunit) and matK (maturase K) are used for barcoding plants. The most abundant protein on earth, RuBisCo catalyzes the first step of carbon fixation, while maturase K encodes a protein that assists RNA editing. A region of the mitochondrial gene COI (cytochrome c oxidase subunit I) is used for barcoding animals. COI is involved in the electron transport phase of respiration. Thus, many genes used for barcoding are involved in the key reactions of life: storing energy in carbohydrates and releasing it to form ATP. COI in fungi and lichens is difficult to amplify, insufficiently variable, and some fungal groups lack mitochondria. Instead, the nuclear internal transcribed spacer (ITS), a variable region that surrounds the 5.8s ribosomal RNA gene, is targeted. Like organelle genes, there are many copies of ITS per genome, and the variability in fungi and lichens allows for their identification. The ITS region is also used for barcoding plants when rbcL and matK do not work. Some organisms need other taxa-specific primers for identification. For instance, green macroalgae lack matKand are difficult to barcode with rbcL and ITS. For these plants, another chloroplast gene, tufA, which codes for elongation factor Tu (EF-Tu), involved in protein synthesis, can be used. DNA barcoding to the species level is sometimes difficult with a single barcode, as species may share identical barcodes. Using multiple barcoding regions can help differentiate these closely related species.
This laboratory uses DNA barcoding to identify plants, fungi, or animals – or products made from them. First, a sample of tissue is collected, preserving the specimen whenever possible and noting its geographical location and local environment. A small leaf disc, a whole insect, or samples of muscle are suitable sources. DNA is extracted from the tissue sample, and the barcode portion of the rbcL, COI and ITS gene is amplified by PCR. The amplified sequence (amplicon) is submitted for sequencing in one or both directions.
The sequencing results are then used to search a DNA database. A close match quickly identifies a species that is already represented in the database. However, some barcodes will be entirely new, and identification may rely on placing the unknown species in a phylogenetic tree with near relatives. Novel DNA barcodes can be submitted to GenBank® (http://www.ncbi.nlm.nih.gov).
- Hebert P.D., Cywinska A., Ball S.L., deWaard J.R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313-21.
- Hebert P.D.N., Penton E.H., Burns J.M., Janzen D.H., Hallwachs W. (2004). Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 101(41):14812-7.
- Hollingsworth P.M. et al (2009). A DNA barcode for land plants. Proc Natl Acad Sci USA 106(31): 12794-7.
- Ratnasingham, S., Hebert, P.D.N (2007). BOLD: The Barcode of Life Data System. Molecular Ecology Notes 7(3): 355-64.
- Stoeckle M. (2003). Taxonomy, DNA, and the Bar Code of Life. BioScience 53(9): 2-3.
- Van Den Berg C., Higgins W.E., Dressler R.L., Whitten W.M., Soto-Arenas M.A., Chase M.W. (2009) A phylogenetic study of Laeliinae (Orchidaceae) based on combined nuclear and plastid DNA sequences. Annals of Botany 104(3): 417-30.
- Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I, Lipman D.J., Ostell J., Sayers E.W. (2013). Nucleic Acids Res. GenBank. 41(D1): D36–D42.

 Download Protocol PDF
Download Protocol PDF